Application on non-model organisms
Current methods allow the reconstruction of high-quality GSMs but do not always take into account the need for metadata storage and exploitation to facilitate the study and reproducibility of models.

We designed four different pipelines for the genome-scale metabolic reconstruction of a brown alga (Ectocarpus siliculosus), a micro-alga (Tisochrysis lutea), and two bacteria (Sulfobacillus thermosulfidooxidans strain Cutipay, and Enterococcus faecalis). GSMs resulting from these pipelines possess the metadata associated with the reconstruction process; enabling the classification of every reaction according to the step that led to its addition in the model, as well as biological metadata. The pipelines vary between the four organisms in terms of gap-filling and analysis tools used.
Learn more about Tisochrysis lutea reconstruction ...
The benefit of tracking process metadata during the combination of orthology, annotation and gap-filling is also noticeable at the pathway scale. Complementary methods that exploit all available data can retrieve several reactions from pathways; resulting in the reconstruction of complete or near exhaustive pathways. An example of pathway completion can be observed during the reconstruction process of E. siliculosus. Having access to traceability metadata allows the user to check the origin of every added reaction of a pathway. Also, it is useful to evaluate the confidence linked to the pathway reactions through the inspection of the presence or absence of associated genes.
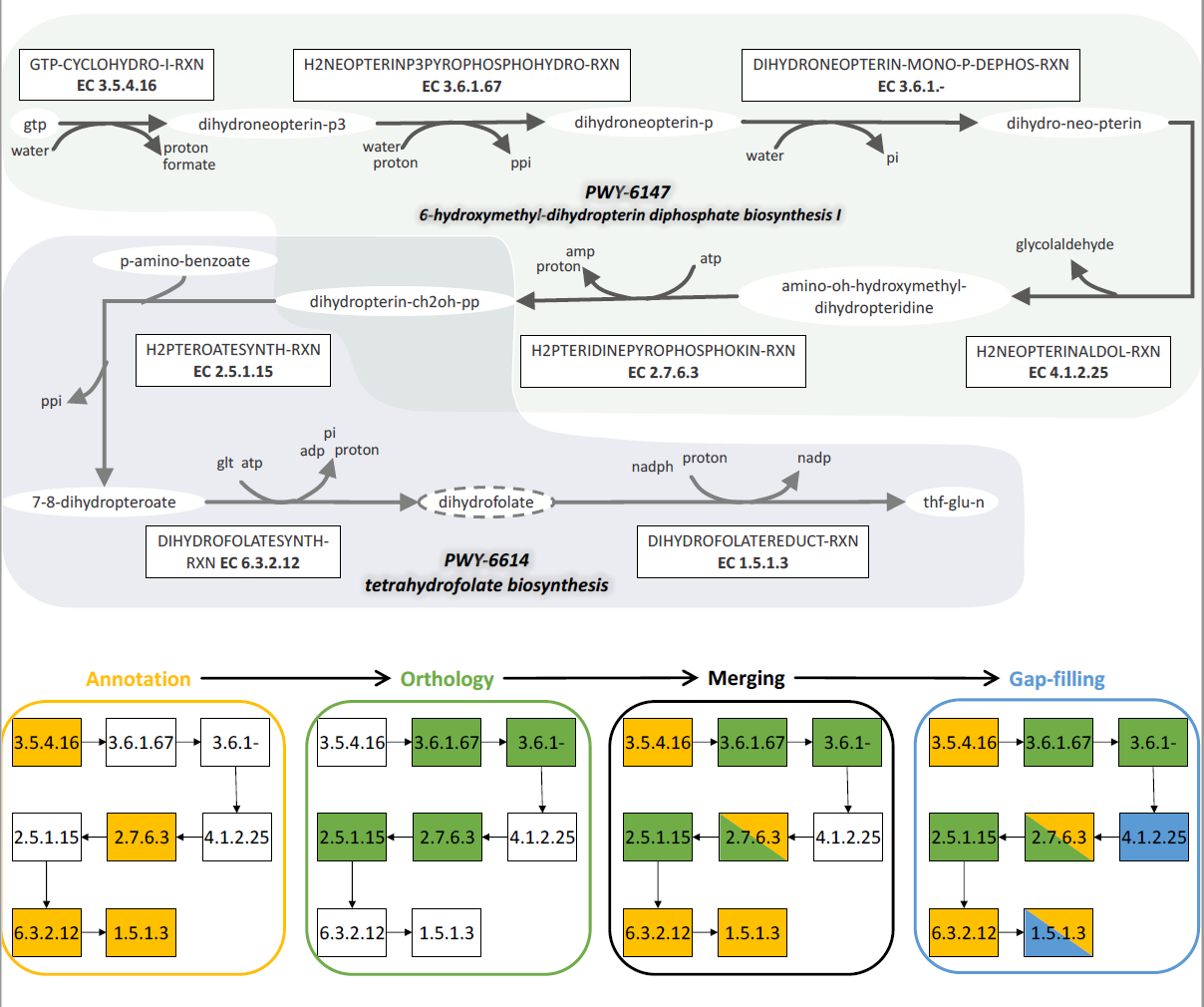
Completion of the 6-hydroxymethyl-dihydropterin diphosphate biosynthesis I and the tetrahydrofolate biosynthesis pathways in E. siliculosus via the combination of annotation (yellow), orthology (green) and gap-filling (blue). The dihydrofolate compound with the dotted line is an instance of the dihydrofolate-glu-n class, following MetaCyc classes ontology structure. The class compound is the original reactant of the dihydrofolatereduct-rxn reaction retrieved with annotation, whereas the previous reaction of the pathway (dihydrofolatesynth-rxn) produces the instance dihydrofolate. Hence the gap-filling step that, using an extended version of MetaCyc, selects an instantiated version of dihydrofolatesynth-rxn that consumes the instance dihydrofolate.

