About PADMet in AuReMe
The AuReMe workspace has many features included. The PADMet package is part of them, enabling users to link admissible input data to the customized workflows and the various analysis tools available in the workspace.
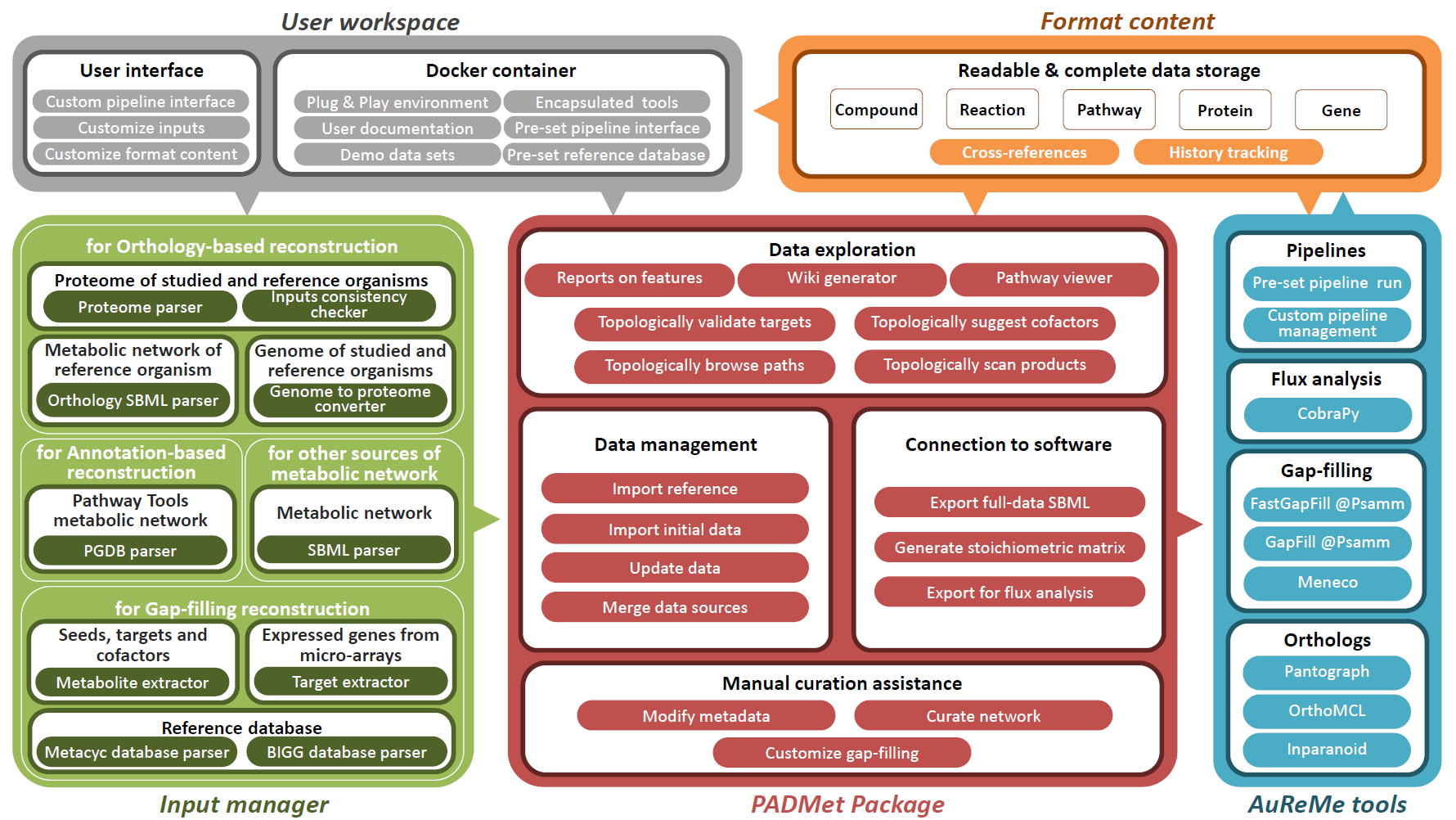
The AuReMe workspace is encapsulated in a Docker image with a “toy” dataset along with versions of MetaCyc 20.5, 22.0 and BiGG 2.3 databases. The purpose of the Docker encapsulation is to provide a workspace that can easily be used with the most diverse set of operating systems (Linux, MacOS, Windows). PADMet format ensures the interoperability of knowledge, tools and data.
Some of the AuReMe's features :- Adaptability to various input data and databases
- Customization of a pre-set pipeline
- Assisted and tracked manual curation
- Opened to other platforms
PADMet-utils (set of tools based on the PADMet package) proposes several solutions for exploring GSMs such as the creation of pathway-completeness text or graphics reports. As a main originality, PADMet-utils allows a user to automatically create a local wiki containing all the information related to the model, including its metadata.
We put online wikis about some metabolic networks reconstructed thanks to AuReMe
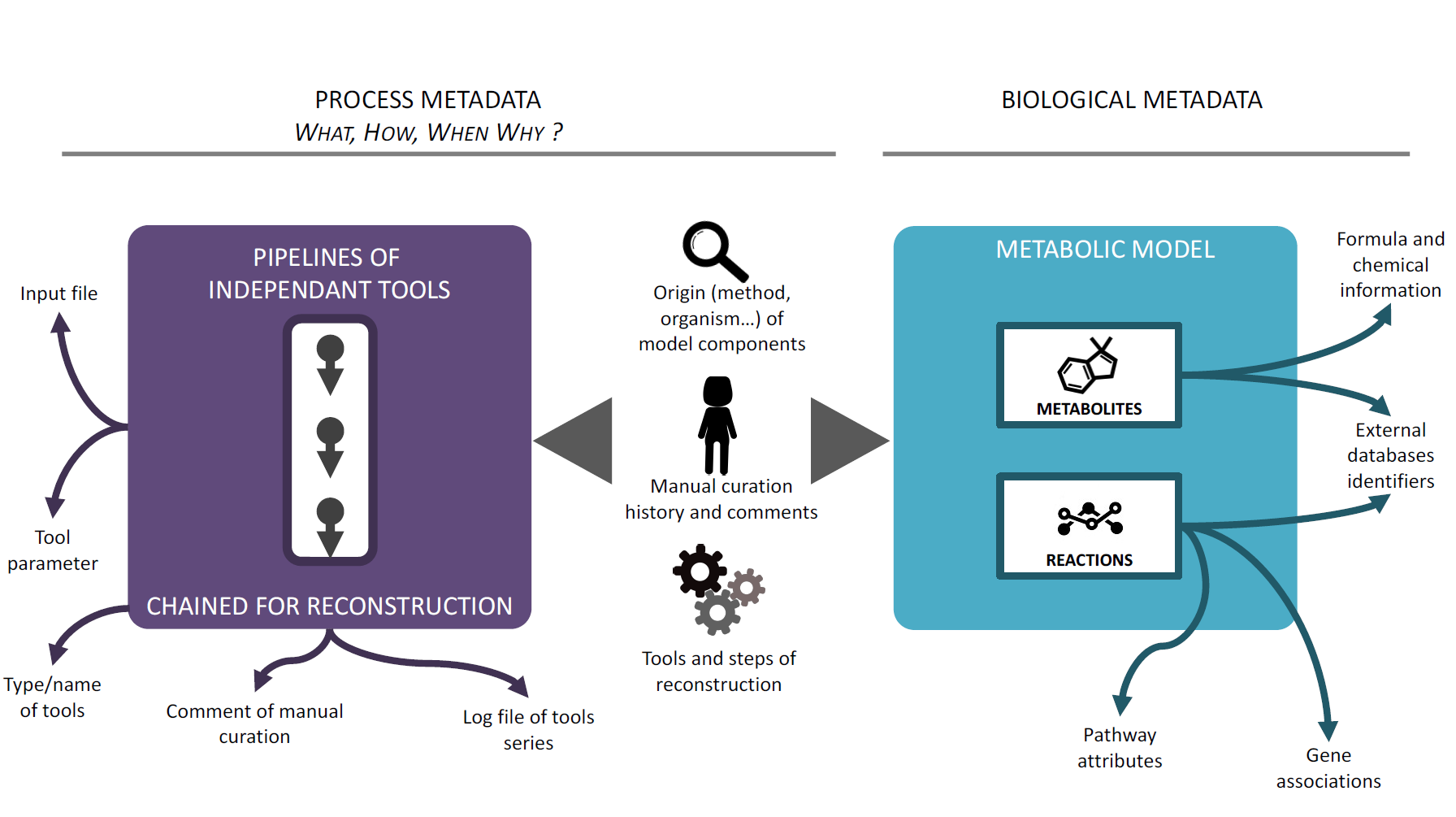
We consider two types of metadata: biological metadata associated ith the metabolic model, and process metadata associated with the reconstruction process.